系统生物学
此词条暂由彩云小译翻译,未经人工整理和审校,带来阅读不便,请见谅。
An illustration of the systems approach to biology
系统方法研究生物学的一个例证
Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionism) to biological research.[1] When it is crossing the field of systems theory and the applied mathematics methods, it develops into the sub-branch of complex systems biology.
Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach (holism instead of the more traditional reductionism) to biological research. When it is crossing the field of systems theory and the applied mathematics methods, it develops into the sub-branch of complex systems biology.
系统生物学是对复杂生物系统进行计算、数学分析和建模的学科。它是一个以生物学为基础的跨学科研究领域,侧重于生物系统内复杂的相互作用,采用整体的方法(整体论而不是更传统的还原论)进行生物学研究。它跨越了系统论和应用数学方法的领域,发展成为复杂系统生物学的一个分支。
Particularly from year 2000 onwards, the concept has been used widely in biology in a variety of contexts. The Human Genome Project is an example of applied systems thinking in biology which has led to new, collaborative ways of working on problems in the biological field of genetics.[2] One of the aims of systems biology is to model and discover emergent properties, properties of cells, tissues and organisms functioning as a system whose theoretical description is only possible using techniques of systems biology.[3][1] These typically involve metabolic networks or cell signaling networks.[4][1]
Particularly from year 2000 onwards, the concept has been used widely in biology in a variety of contexts. The Human Genome Project is an example of applied systems thinking in biology which has led to new, collaborative ways of working on problems in the biological field of genetics. One of the aims of systems biology is to model and discover emergent properties, properties of cells, tissues and organisms functioning as a system whose theoretical description is only possible using techniques of systems biology.
特别是从2000年起,这个概念在生物学中被广泛应用于各种场合。人类基因组计划是生物学中应用系统思维的一个例子,它导致了在遗传学这个生物学领域新的协作的工作方式。系统生物学的目标之一是模拟和发现细胞、组织和有机体作为一个系统运作的涌现特性,其理论描述只有使用系统生物学技术才有可能实现。
Overview
Systems biology can be considered from a number of different aspects.
Systems biology can be considered from a number of different aspects.
系统生物学可以从许多不同的方面来考虑。
As a field of study, particularly, the study of the interactions between the components of biological systems, and how these interactions give rise to the function and behavior of that system (for example, the enzymes and metabolites in a metabolic pathway or the heart beats).[5][6][7]
As a field of study, particularly, the study of the interactions between the components of biological systems, and how these interactions give rise to the function and behavior of that system (for example, the enzymes and metabolites in a metabolic pathway or the heart beats).
系统生物学是这样一个研究领域,研究生物系统各组成部分之间的相互作用,以及这些相互作用如何产生该系统的功能和行为(例如,代谢通路或心跳中的酶和代谢物)。
As a paradigm, systems biology is usually defined in antithesis to the so-called reductionist paradigm (biological organisation), although it's fully consistent with the scientific method. The distinction between the two paradigms is referred to in these quotations: "The reductionist approach has successfully identified most of the components and many of the interactions but, unfortunately, offers no convincing concepts or methods to understand how system properties emerge ... the pluralism of causes and effects in biological networks is better addressed by observing, through quantitative measures, multiple components simultaneously and by rigorous data integration with mathematical models." (Sauer et al.)[8] "Systems biology ... is about putting together rather than taking apart, integration rather than reduction. It requires that we develop ways of thinking about integration that are as rigorous as our reductionist programmes, but different. ... It means changing our philosophy, in the full sense of the term." (Denis Noble)[7]
As a paradigm, systems biology is usually defined in antithesis to the so-called reductionist paradigm (biological organisation), although it's fully consistent with the scientific method. The distinction between the two paradigms is referred to in these quotations: "The reductionist approach has successfully identified most of the components and many of the interactions but, unfortunately, offers no convincing concepts or methods to understand how system properties emerge ... the pluralism of causes and effects in biological networks is better addressed by observing, through quantitative measures, multiple components simultaneously and by rigorous data integration with mathematical models." (Sauer et al.) "Systems biology ... is about putting together rather than taking apart, integration rather than reduction. It requires that we develop ways of thinking about integration that are as rigorous as our reductionist programmes, but different. ... It means changing our philosophy, in the full sense of the term." (Denis Noble)
作为一种研究范式,系统生物学通常被认为与所谓的还原论范式(生物组织)相对立,尽管它完全符合科学方法。这些语录中提到了两种范式之间的区别: “还原论方法成功地确定了大多数组成部分和许多相互作用,但不幸的是,没有提供令人信服的概念或方法来理解系统特性是如何出现的... 生物网络中因果关系的多元性最好通过多个组分同时进行定量测量,以及与数学模型进行严格的数据整合来解决。”(Sauer 等人)“系统生物学...是关于合并而不是分解,是关于整合而不是简化。它要求我们发展出与我们的还原论方法一样严谨但不同的整合思维方式...这意味着彻底改变我们的哲学。”(丹尼斯 · 诺贝尔)
As a series of operational protocols used for performing research, namely a cycle composed of theory, analytic or computational modelling to propose specific testable hypotheses about a biological system, experimental validation, and then using the newly acquired quantitative description of cells or cell processes to refine the computational model or theory.[9] Since the objective is a model of the interactions in a system, the experimental techniques that most suit systems biology are those that are system-wide and attempt to be as complete as possible. Therefore, transcriptomics, metabolomics, proteomics and high-throughput techniques are used to collect quantitative data for the construction and validation of models.[10]
As a series of operational protocols used for performing research, namely a cycle composed of theory, analytic or computational modelling to propose specific testable hypotheses about a biological system, experimental validation, and then using the newly acquired quantitative description of cells or cell processes to refine the computational model or theory. Since the objective is a model of the interactions in a system, the experimental techniques that most suit systems biology are those that are system-wide and attempt to be as complete as possible. Therefore, transcriptomics, metabolomics, proteomics and high-throughput techniques are used to collect quantitative data for the construction and validation of models.
系统生物学是一系列用于进行研究的操作方案,即一个由理论、分析或计算模型组成的循环,首先提出关于生物系统的具体可检验的假设,接着进行实验验证,然后使用新获得的细胞或细胞过程的定量描述来优化计算模型或理论。既然目标是一个系统中相互作用的模型,那么最适合系统生物学的实验技术就是那些全系统范围的、尽可能完整的实验技术。因此,转录组学、代谢组学、蛋白质组学和高通量技术被用来收集定量数据,从而用于模型的建立和验证。
As the application of dynamical systems theory to molecular biology. Indeed, the focus on the dynamics of the studied systems is the main conceptual difference between systems biology and bioinformatics.[11]
As the application of dynamical systems theory to molecular biology. Indeed, the focus on the dynamics of the studied systems is the main conceptual difference between systems biology and bioinformatics.
系统生物学是动力系统理论在分子生物学中的应用。事实上,对所研究系统的动力学的关注是系统生物学和生物信息学之间的主要概念差异。
As a socioscientific phenomenon defined by the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources using interdisciplinary tools and personnel.[12]
As a socioscientific phenomenon defined by the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources using interdisciplinary tools and personnel.
系统生物学也是一种社会科学现象,它利用跨学科的工具和人员,从不同的实验来源整合有关生物系统相互作用的复杂数据。
This variety of viewpoints is illustrative of the fact that systems biology refers to a cluster of peripherally overlapping concepts rather than a single well-delineated field. However, the term has widespread currency and popularity as of 2007, with chairs and institutes of systems biology proliferating worldwide.
This variety of viewpoints is illustrative of the fact that systems biology refers to a cluster of peripherally overlapping concepts rather than a single well-delineated field. However, the term has widespread currency and popularity as of 2007, with chairs and institutes of systems biology proliferating worldwide.
各种各样的观点说明了这样一个事实,即系统生物学指的是一系列周边重叠概念的集合,而不是一个单独界定清楚的领域。然而,随着系统生物学的教职和研究机构在全球范围内的激增,这个术语在2007年已经广泛流行和普及。
History
Systems biology finds its roots in[citation needed] the quantitative modeling of enzyme kinetics, a discipline that flourished between 1900 and 1970, the mathematical modeling of population dynamics, the simulations developed to study neurophysiology, control theory and cybernetics, and synergetics.
Systems biology finds its roots in the quantitative modeling of enzyme kinetics, a discipline that flourished between 1900 and 1970, the mathematical modeling of population dynamics, the simulations developed to study neurophysiology, control theory and cybernetics, and synergetics.
系统生物学根植于酶动力学的定量模型,这门学科在1900年到1970年间蓬勃发展,族群动态的数学模型,为研究神经生理学、控制理论和控制论以及协同学而开发的模拟。
One of the theorists who can be seen as one of the precursors of systems biology is Ludwig von Bertalanffy with his general systems theory.[13] One of the first numerical simulations in cell biology was published in 1952 by the British neurophysiologists and Nobel prize winners Alan Lloyd Hodgkin and Andrew Fielding Huxley, who constructed a mathematical model that explained the action potential propagating along the axon of a neuronal cell.[14] Their model described a cellular function emerging from the interaction between two different molecular components, a potassium and a sodium channel, and can therefore be seen as the beginning of computational systems biology.[15] Also in 1952, Alan Turing published The Chemical Basis of Morphogenesis, describing how non-uniformity could arise in an initially homogeneous biological system.[16]
One of the theorists who can be seen as one of the precursors of systems biology is Ludwig von Bertalanffy with his general systems theory.
其中一个可以被看作是系统生物学先驱的理论家用他的一般系统理论卡尔·路德维希·冯·贝塔郎非。
In 1960, Denis Noble developed the first computer model of the heart pacemaker.[17]
In 1960, Denis Noble developed the first computer model of the heart pacemaker.
1960年,丹尼斯 · 诺布尔发明了第一个心脏起搏器的计算机模型。
The formal study of systems biology, as a distinct discipline, was launched by systems theorist Mihajlo Mesarovic in 1966 with an international symposium at the Case Institute of Technology in Cleveland, Ohio, titled "Systems Theory and Biology".[18][19]
The formal study of systems biology, as a distinct discipline, was launched by systems theorist Mihajlo Mesarovic in 1966 with an international symposium at the Case Institute of Technology in Cleveland, Ohio, titled "Systems Theory and Biology".
系统生物学是一门独特的学科,系统理论家米哈伊洛 · 梅萨罗维奇于1966年在俄亥俄州克利夫兰市的凯斯理工学院召开了一次题为“系统理论与生物学”的国际研讨会,正式开展了系统生物学的研究。
The 1960s and 1970s saw the development of several approaches to study complex molecular systems, such as the metabolic control analysis and the biochemical systems theory. The successes of molecular biology throughout the 1980s, coupled with a skepticism toward theoretical biology, that then promised more than it achieved, caused the quantitative modeling of biological processes to become a somewhat minor field.[20]
The 1960s and 1970s saw the development of several approaches to study complex molecular systems, such as the metabolic control analysis and the biochemical systems theory. The successes of molecular biology throughout the 1980s, coupled with a skepticism toward theoretical biology, that then promised more than it achieved, caused the quantitative modeling of biological processes to become a somewhat minor field.
20世纪60年代和70年代发展了几种研究复杂分子系统的方法,如代谢控制分析和生化系统理论。整个20世纪80年代分子生物学的成功,加上对理论生物学的怀疑,使得生物过程的定量模拟成为一个有点次要的领域。
Shows trends in systems biology research by presenting the number of articles out of the top 30 cited systems biology papers during that time which include a specific topic
显示系统生物学研究的趋势,通过提出的文章数量前30个被引用的系统生物学论文在这段时间内,其中包括一个具体的主题
However, the birth of functional genomics in the 1990s meant that large quantities of high-quality data became available, while the computing power exploded, making more realistic models possible. In 1992, then 1994, serial articles 引用错误:没有找到与</ref>对应的<ref>标签[22][23][24][25][26][27] on systems medicine, systems genetics, and systems biological engineering by B. J. Zeng was published in China and was giving a lecture on biosystems theory and systems-approach research at the First International Conference on Transgenic Animals, Beijing, 1996. In 1997, the group of Masaru Tomita published the first quantitative model of the metabolism of a whole (hypothetical) cell.[28]
No.8-10, 1996. Etc.</ref> on systems medicine, systems genetics, and systems biological engineering by B. J. Zeng was published in China and was giving a lecture on biosystems theory and systems-approach research at the First International Conference on Transgenic Animals, Beijing, 1996. In 1997, the group of Masaru Tomita published the first quantitative model of the metabolism of a whole (hypothetical) cell.
1996年8-10号。系统医学、系统遗传学和系统生物工程学等。曾斌于1996年在中国出版了《系统医学、系统遗传学和系统生物工程》 ,并在北京举行的第一届国际转基因动物会议上作了关于生物系统理论和系统方法研究的演讲。1997年,富田正丸小组发表了第一个关于整个(假设的)细胞新陈代谢的定量模型。
Around the year 2000, after Institutes of Systems Biology were established in Seattle and Tokyo, systems biology emerged as a movement in its own right, spurred on by the completion of various genome projects, the large increase in data from the omics (e.g., genomics and proteomics) and the accompanying advances in high-throughput experiments and bioinformatics. Shortly afterwards, the first departments wholly devoted to systems biology were founded (for example, the Department of Systems Biology at Harvard Medical School [29]).
Around the year 2000, after Institutes of Systems Biology were established in Seattle and Tokyo, systems biology emerged as a movement in its own right, spurred on by the completion of various genome projects, the large increase in data from the omics (e.g., genomics and proteomics) and the accompanying advances in high-throughput experiments and bioinformatics. Shortly afterwards, the first departments wholly devoted to systems biology were founded (for example, the Department of Systems Biology at Harvard Medical School ).
2000年左右,在西雅图和东京建立了系统生物学研究所之后,由于各种基因组项目的完成、组学(如基因组学和蛋白质组学)数据的大量增加以及随之而来的高通量实验和生物信息学的进展,系统生物学作为一项独立的运动而出现。不久之后,第一个完全致力于系统生物学的系成立了(例如,哈佛医学院的系统生物学系)。
In 2003, work at the Massachusetts Institute of Technology was begun on CytoSolve, a method to model the whole cell by dynamically integrating multiple molecular pathway models.[30] Since then, various research institutes dedicated to systems biology have been developed. For example, the NIGMS of NIH established a project grant that is currently supporting over ten systems biology centers in the United States.[31] As of summer 2006, due to a shortage of people in systems biology[32] several doctoral training programs in systems biology have been established in many parts of the world. In that same year, the National Science Foundation (NSF) put forward a grand challenge for systems biology in the 21st century to build a mathematical model of the whole cell.[citation needed] In 2012 the first whole-cell model of Mycoplasma genitalium was achieved by the Karr Laboratory at the Mount Sinai School of Medicine in New York. The whole-cell model is able to predict viability of M. genitalium cells in response to genetic mutations.[33]
In 2003, work at the Massachusetts Institute of Technology was begun on CytoSolve, a method to model the whole cell by dynamically integrating multiple molecular pathway models. Since then, various research institutes dedicated to systems biology have been developed. For example, the NIGMS of NIH established a project grant that is currently supporting over ten systems biology centers in the United States. As of summer 2006, due to a shortage of people in systems biology
2003年,麻省理工学院开始研究细胞解决方案,这是一种通过动态整合多个分子通路模型来建立整个细胞模型的方法。从那时起,各种致力于系统生物学的研究机构已经发展起来。例如,美国国立卫生研究院的 NIGMS 建立了一个项目补助金,目前正在支持美国的十多个系统生物学中心。截至2006年夏天,由于系统生物学人才短缺
An important milestone in the development of systems biology has become the international project Physiome.
An important milestone in the development of systems biology has become the international project Physiome.
系统生物学发展的一个重要里程碑已经成为国际性的物理学课题。
Associated disciplines
Overview of signal transduction pathways
[信号转导/出路]概览
According to the interpretation of Systems Biology as the ability to obtain, integrate and analyze complex data sets from multiple experimental sources using interdisciplinary tools, some typical technology platforms are phenomics, organismal variation in phenotype as it changes during its life span; genomics, organismal deoxyribonucleic acid (DNA) sequence, including intra-organismal cell specific variation. (i.e., telomere length variation); epigenomics/epigenetics, organismal and corresponding cell specific transcriptomic regulating factors not empirically coded in the genomic sequence. (i.e., DNA methylation, Histone acetylation and deacetylation, etc.); transcriptomics, organismal, tissue or whole cell gene expression measurements by DNA microarrays or serial analysis of gene expression; interferomics, organismal, tissue, or cell-level transcript correcting factors (i.e., RNA interference), proteomics, organismal, tissue, or cell level measurements of proteins and peptides via two-dimensional gel electrophoresis, mass spectrometry or multi-dimensional protein identification techniques (advanced HPLC systems coupled with mass spectrometry). Sub disciplines include phosphoproteomics, glycoproteomics and other methods to detect chemically modified proteins; metabolomics, measurements of small molecules known as metabolites in the system at the organismal, cell, or tissue level;[34] glycomics, organismal, tissue, or cell-level measurements of carbohydrates; lipidomics, organismal, tissue, or cell level measurements of lipids.
According to the interpretation of Systems Biology as the ability to obtain, integrate and analyze complex data sets from multiple experimental sources using interdisciplinary tools, some typical technology platforms are phenomics, organismal variation in phenotype as it changes during its life span; genomics, organismal deoxyribonucleic acid (DNA) sequence, including intra-organismal cell specific variation. (i.e., telomere length variation); epigenomics/epigenetics, organismal and corresponding cell specific transcriptomic regulating factors not empirically coded in the genomic sequence. (i.e., DNA methylation, Histone acetylation and deacetylation, etc.); transcriptomics, organismal, tissue or whole cell gene expression measurements by DNA microarrays or serial analysis of gene expression; interferomics, organismal, tissue, or cell-level transcript correcting factors (i.e., RNA interference), proteomics, organismal, tissue, or cell level measurements of proteins and peptides via two-dimensional gel electrophoresis, mass spectrometry or multi-dimensional protein identification techniques (advanced HPLC systems coupled with mass spectrometry). Sub disciplines include phosphoproteomics, glycoproteomics and other methods to detect chemically modified proteins; metabolomics, measurements of small molecules known as metabolites in the system at the organismal, cell, or tissue level; glycomics, organismal, tissue, or cell-level measurements of carbohydrates; lipidomics, organismal, tissue, or cell level measurements of lipids.
根据系统生物学的解释,即利用跨学科工具从多个实验来源获取、整合和分析复杂数据集的能力,一些典型的技术平台包括表型学、生物表型在其生命周期内发生变化时的变异、基因组学、生物脱氧核糖核酸序列,包括生物内部细胞特异性变异。表观基因组学/表观遗传学、有机体和相应的细胞特异性转录调控因子,这些因子没有经验性地编码在基因组序列中。DNA 甲基化、组蛋白乙酰化和脱乙酰化等。) ; 转录组、生物组织、组织或整个细胞的基因表达测量,通过 DNA 微阵列或基因表达的系列分析; 干扰物、生物组织、组织或细胞水平的转录校正因子(即 RNA干扰) ; 蛋白质组学、生物组织、组织或细胞水平的蛋白质和多肽测量,通过二维凝胶电泳、质谱法或多维蛋白质识别技术(先进的高效液相色谱系统加上质谱法)。子学科包括磷酸蛋白质组学、糖蛋白质组学和其他检测化学修饰蛋白质的方法; 代谢组学,测量有机体、细胞或组织水平系统中被称为代谢物的小分子; 糖组学、有机体、组织或细胞水平的碳水化合物测量; 脂质组学、有机体、组织或细胞水平的脂质测量。
In addition to the identification and quantification of the above given molecules further techniques analyze the dynamics and interactions within a cell.The interactions studied include organismal, tissue, cell, and molecular interactions within the cell (interactomics).[35] Currently, the authoritative molecular discipline in this field of study is protein-protein interactions (PPI), although the working definition does not preclude inclusion of other molecular disciplines. These molecular disciplines include; neuroelectrodynamics, an organismal network where the brain's computing function as a dynamic system includes underlying biophysical mechanisms and emerging computation by electrical interactions;[36] fluxomics, measurements of molecular dynamic changes over time in a system such as a cell, tissue, or organism;[34] biomics, systems analysis of the biome; and molecular biokinematics, the study of "biology in motion" focused on how cells transit between steady states such as in proteins molecular mechanism.[37]
In addition to the identification and quantification of the above given molecules further techniques analyze the dynamics and interactions within a cell.The interactions studied include organismal, tissue, cell, and molecular interactions within the cell (interactomics). Currently, the authoritative molecular discipline in this field of study is protein-protein interactions (PPI), although the working definition does not preclude inclusion of other molecular disciplines. These molecular disciplines include; neuroelectrodynamics, an organismal network where the brain's computing function as a dynamic system includes underlying biophysical mechanisms and emerging computation by electrical interactions; fluxomics, measurements of molecular dynamic changes over time in a system such as a cell, tissue, or organism;
除了识别和定量上述给定的分子之外,进一步的技术分析细胞内的动力学和相互作用。研究的相互作用包括生物、组织、细胞和分子在细胞内的相互作用(相互作用)。目前,在这一领域的权威分子学科是蛋白质-蛋白质相互作用(PPI) ,虽然工作的定义并不排除包括其他分子学科。这些分子学科包括: 神经电动力学,这是一个有机体网络,其中大脑的计算功能作为一个动态系统,包括潜在的生物物理机制和新兴的电力相互作用的计算; 流体学,测量分子随着时间的动态变化在一个系统,如细胞,组织,或有机体;
In approaching a systems biology problem there are two main approaches. These are the top down and bottom up approach. The top down approach takes as much of the system into account as possible and relies largely on experimental results. The RNA-seq technique is an example of an experimental top down approach. Conversely, the bottom up approach is used to create detailed models while also incorporating experimental data. An example of the bottom up approach is the use of circuit models to describe a simple gene network.[38]
In approaching a systems biology problem there are two main approaches. These are the top down and bottom up approach. The top down approach takes as much of the system into account as possible and relies largely on experimental results. The RNA-seq technique is an example of an experimental top down approach. Conversely, the bottom up approach is used to create detailed models while also incorporating experimental data. An example of the bottom up approach is the use of circuit models to describe a simple gene network.
在处理系统生物学问题时,有两种主要的方法。这些是自上而下和自下而上的方法。自上而下的方法尽可能多地考虑系统,并且在很大程度上依赖于实验结果。RNA-seq 技术是自顶向下实验方法的一个例子。相反,自底向上的方法用于创建详细的模型,同时也合并了实验数据。自底向上方法的一个例子是使用电路模型来描述一个简单的基因网络。
Various technologies utilized to capture dynamic changes in mRNA, proteins, and post-translational modifications. Mechanobiology, forces and physical properties at all scales, their interplay with other regulatory mechanisms;[39] biosemiotics, analysis of the system of sign relations of an organism or other biosystems; Physiomics, a systematic study of physiome in biology.
Various technologies utilized to capture dynamic changes in mRNA, proteins, and post-translational modifications. Mechanobiology, forces and physical properties at all scales, their interplay with other regulatory mechanisms; biosemiotics, analysis of the system of sign relations of an organism or other biosystems; Physiomics, a systematic study of physiome in biology.
各种技术用于捕获动态变化的信使核糖核酸,蛋白质,和翻译后修饰。机械生物学,所有尺度的力和物理性质,它们与其他调节机制的相互作用; 生物符号学,分析有机体或其他生物系统的符号关系系统; 生理组学,生物学中生理组的系统研究。
Cancer systems biology is an example of the systems biology approach, which can be distinguished by the specific object of study (tumorigenesis and treatment of cancer). It works with the specific data (patient samples, high-throughput data with particular attention to characterizing cancer genome in patient tumour samples) and tools (immortalized cancer cell lines, mouse models of tumorigenesis, xenograft models, high-throughput sequencing methods, siRNA-based gene knocking down high-throughput screenings, computational modeling of the consequences of somatic mutations and genome instability).[40] The long-term objective of the systems biology of cancer is ability to better diagnose cancer, classify it and better predict the outcome of a suggested treatment, which is a basis for personalized cancer medicine and virtual cancer patient in more distant prospective. Significant efforts in computational systems biology of cancer have been made in creating realistic multi-scale in silico models of various tumours.引用错误:没有找到与</ref>对应的<ref>标签
|year=2010|journal=Nature Reviews Cancer|volume=10|issue=3|pages=221–230|pmid=20179714|doi=10.1038/nrc2808|title=Dissecting cancer through mathematics: from the cell to the animal model }}</ref>
| year = 2010 | journal = Nature Reviews Cancer | volume = 10 | issue = 3 | pages = 221-230 | pmd = 20179714 | doi = 10.1038/nrc2808 | title = 通过数学解剖癌症: 从细胞到动物模型} </ref >
The investigations are frequently combined with large-scale perturbation methods, including gene-based (RNAi, mis-expression of wild type and mutant genes) and chemical approaches using small molecule libraries.[citation needed] Robots and automated sensors enable such large-scale experimentation and data acquisition. These technologies are still emerging and many face problems that the larger the quantity of data produced, the lower the quality.[citation needed] A wide variety of quantitative scientists (computational biologists, statisticians, mathematicians, computer scientists and physicists) are working to improve the quality of these approaches and to create, refine, and retest the models to accurately reflect observations.
The investigations are frequently combined with large-scale perturbation methods, including gene-based (RNAi, mis-expression of wild type and mutant genes) and chemical approaches using small molecule libraries. Robots and automated sensors enable such large-scale experimentation and data acquisition. These technologies are still emerging and many face problems that the larger the quantity of data produced, the lower the quality. A wide variety of quantitative scientists (computational biologists, statisticians, mathematicians, computer scientists and physicists) are working to improve the quality of these approaches and to create, refine, and retest the models to accurately reflect observations.
这些研究经常与大规模的微扰方法结合,包括基于基因的(rna 干扰,野生型和突变型基因的错误表达)和使用小分子文库的化学方法。机器人和自动化传感器使这种大规模的实验和数据采集成为可能。这些技术仍在出现,许多面临的问题是,产生的数据量越大,质量就越低。各种各样的定量科学家(计算生物学家、统计学家、数学家、计算机科学家和物理学家)正在努力提高这些方法的质量,并创建、完善和重新测试模型,以准确地反映观测结果。
The systems biology approach often involves the development of mechanistic models, such as the reconstruction of dynamic systems from the quantitative properties of their elementary building blocks.[41][42][43][44] For instance, a cellular network can be modelled mathematically using methods coming from chemical kinetics[45] and control theory. Due to the large number of parameters, variables and constraints in cellular networks, numerical and computational techniques are often used (e.g., flux balance analysis).[43][45]
The systems biology approach often involves the development of mechanistic models, such as the reconstruction of dynamic systems from the quantitative properties of their elementary building blocks. For instance, a cellular network can be modelled mathematically using methods coming from chemical kinetics and control theory. Due to the large number of parameters, variables and constraints in cellular networks, numerical and computational techniques are often used (e.g., flux balance analysis).
系统生物学方法经常涉及机械模型的发展,例如从动态系统的基本构件的定量特性重建动态系统。例如,一个蜂窝网络可以用数学方法来建模,使用的方法来自化学动力学和控制理论。由于蜂窝网络中参数、变量和约束的数量庞大,经常使用数值和计算技术(例如通量平衡分析)。
Bioinformatics and data analysis
Other aspects of computer science, informatics, and statistics are also used in systems biology. These include new forms of computational models, such as the use of process calculi to model biological processes (notable approaches include stochastic π-calculus, BioAmbients, Beta Binders, BioPEPA, and Brane calculus) and constraint-based modeling; integration of information from the literature, using techniques of information extraction and text mining;[46] development of online databases and repositories for sharing data and models, approaches to database integration and software interoperability via loose coupling of software, websites and databases, or commercial suits; network-based approaches for analyzing high dimensional genomic data sets. For example, weighted correlation network analysis is often used for identifying clusters (referred to as modules), modeling the relationship between clusters, calculating fuzzy measures of cluster (module) membership, identifying intramodular hubs, and for studying cluster preservation in other data sets; pathway-based methods for omics data analysis, e.g. approaches to identify and score pathways with differential activity of their gene, protein, or metabolite members.[47] Much of the analysis of genomic data sets also include identifying correlations. Additionally, as much of the information comes from different fields, the development of syntactically and semantically sound ways of representing biological models is needed.[48]
Other aspects of computer science, informatics, and statistics are also used in systems biology. These include new forms of computational models, such as the use of process calculi to model biological processes (notable approaches include stochastic π-calculus, BioAmbients, Beta Binders, BioPEPA, and Brane calculus) and constraint-based modeling; integration of information from the literature, using techniques of information extraction and text mining; development of online databases and repositories for sharing data and models, approaches to database integration and software interoperability via loose coupling of software, websites and databases, or commercial suits; network-based approaches for analyzing high dimensional genomic data sets. For example, weighted correlation network analysis is often used for identifying clusters (referred to as modules), modeling the relationship between clusters, calculating fuzzy measures of cluster (module) membership, identifying intramodular hubs, and for studying cluster preservation in other data sets; pathway-based methods for omics data analysis, e.g. approaches to identify and score pathways with differential activity of their gene, protein, or metabolite members. Much of the analysis of genomic data sets also include identifying correlations. Additionally, as much of the information comes from different fields, the development of syntactically and semantically sound ways of representing biological models is needed.
计算机科学、信息学和统计学的其他方面也用于系统生物学。这些包括新形式的计算模型,如使用过程计算模型生物过程(著名的方法包括随机演算,BioAmbients,Beta Binders,BioPEPA 和 Brane 演算)和基于约束的建模; 综合来自文献的信息,使用信息抽取和文本挖掘技术; 开发在线数据库和存储库共享数据和模型,数据库集成方法和软件互操作性通过松散耦合的软件,网站和数据库,或商业诉讼; 基于网络的方法分析高维基因组数据集。例如,加权相关网络分析常常用于识别集群(称为模块)、建立集群之间的关系模型、计算集群(模块)隶属度的模糊度量、识别模块内集线器,以及用于研究其他数据集中的集群保存; 基于路径的组学数据分析方法,例如。识别和评分不同活性的基因、蛋白质或代谢物成员的途径的方法。许多基因组数据集的分析也包括确定相关性。此外,由于大量的信息来自不同的领域,发展的语法和语义健全的方式表示生物模型是必要的。
Creating biological models
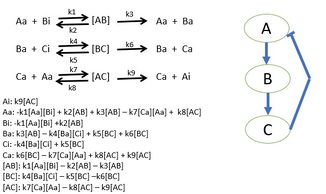
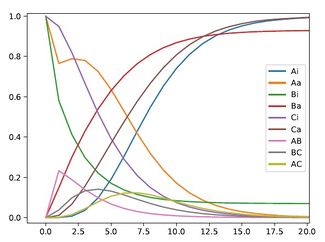
A simple three protein negative feedback loop modeled with mass action kinetic differential equations. Each protein interaction is described by a Michealis Menten reaction.
用质量作用动力学微分方程建立简单的三蛋白质负反馈回路。每个蛋白质的相互作用都是通过米氏反应来描述的。
Researchers begin by choosing a biological pathway and diagramming all of the protein interactions. After determining all of the interactions of the proteins, mass action kinetics is utilized to describe the speed of the reactions in the system. Mass action kinetics will provide differential equations to model the biological system as a mathematical model in which experiments can determine the parameter values to use in the differential equations.[50] These parameter values will be the reaction rates of each proteins interaction in the system. This model determines the behavior of certain proteins in biological systems and bring new insight to the specific activities of individual proteins. Sometimes it is not possible to gather all reaction rates of a system. Unknown reaction rates are determined by simulating the model of known parameters and target behavior which provides possible parameter values.[51][49]
Researchers begin by choosing a biological pathway and diagramming all of the protein interactions. After determining all of the interactions of the proteins, mass action kinetics is utilized to describe the speed of the reactions in the system. Mass action kinetics will provide differential equations to model the biological system as a mathematical model in which experiments can determine the parameter values to use in the differential equations. These parameter values will be the reaction rates of each proteins interaction in the system. This model determines the behavior of certain proteins in biological systems and bring new insight to the specific activities of individual proteins. Sometimes it is not possible to gather all reaction rates of a system. Unknown reaction rates are determined by simulating the model of known parameters and target behavior which provides possible parameter values.
研究人员首先选择一条生物通路,绘制所有蛋白质相互作用的图表。在确定了所有的相互作用的蛋白质,质量作用动力学是用来描述的速度反应的系统。质量作用动力学将提供微分方程来模拟生物系统作为一个数学模型,在这个数学模型中,实验可以确定用于微分方程的参数值。这些参数值将是系统中每个蛋白质相互作用的反应速率。这个模型决定了某些蛋白质在生物系统中的行为,并为单个蛋白质的特定活动带来了新的洞察力。有时不可能收集一个系统的所有反应速率。通过模拟已知参数的模型和提供可能参数值的目标行为,确定未知反应速率。
Plot of Concentrations vs time for the simple three protein negative feedback loop. All parameters are set to either 0 or 1 for initial conditions. The reaction is allowed to proceed until it hits equilibrium. This plot is of the change in each protein over time.
简单的三蛋白质负反馈回路的浓度与时间图。对于初始条件,所有参数设置为0或1。反应继续进行,直到达到平衡。这张图是每个蛋白质随时间的变化。
See also
References
<references>}}
Further reading
- Klipp, Edda; Liebermeister, Wolfram; Wierling, Christoph; Kowald, Axel (2016). Systems Biology - A Textbook, 2nd edition. Wiley. ISBN 978-3-527-33636-4.
- Asfar S. Azmi, ed. (2012). Systems Biology in Cancer Research and Drug Discovery. ISBN 978-94-007-4819-4.
- Kitano, Hiroaki (15 October 2001). Foundations of Systems Biology. MIT Press. ISBN 978-0-262-11266-6. https://archive.org/details/foundationsofsys00hiro.
- Werner, Eric (29 March 2007). "All systems go". Nature. 446 (7135): 493–494. Bibcode:2007Natur.446..493W. doi:10.1038/446493a. provides a comparative review of three books:
- Alon, Uri (7 July 2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall. ISBN 978-1-58488-642-6.
- Kaneko, Kunihiko (15 September 2006). Life: An Introduction to Complex Systems Biology. Springer-Verlag. Bibcode 2006lics.book.....K. ISBN 978-3-540-32666-3.
- Palsson, Bernhard O. (16 January 2006). Systems Biology: Properties of Reconstructed Networks. Cambridge University Press. ISBN 978-0-521-85903-5.
- Encyclopedia of Systems Biology. Springer-Verlag. 13 August 2013. ISBN 978-1-4419-9864-4.
External links
| 40x40px | Look up 系统生物学 in Wiktionary, the free dictionary. |
Category:Bioinformatics
类别: 生物信息学
Category:Computational fields of study
类别: 研究的计算领域
This page was moved from wikipedia:en:Systems biology. Its edit history can be viewed at 系统生物学/edithistory
- 有参考文献错误的页面
- 调用重复模板参数的页面
- CS1 English-language sources (en)
- Articles with short description
- All articles with unsourced statements
- Articles with unsourced statements from May 2009
- Articles with invalid date parameter in template
- 含有受损文件链接的页面
- Articles with unsourced statements from October 2019
- AC with 0 elements
- Pages with red-linked authority control categories
- Systems biology
- Bioinformatics
- Computational fields of study
- 待整理页面